LGCM Tutorial
This tutorial walks through a complete LGCM analysis. By the end, you will have:
- Simulated longitudinal data with known parameters
- Visualized individual trajectories
- Fit and compared growth models
- Interpreted the results
Workflow Overview
┌─────────────────────────────────┐
│ 1. Setup │
│ Load packages, set seed │
└────────────────┬────────────────┘
▼
┌─────────────────────────────────┐
│ 2. Simulate/Load Data │
│ Wide format, check structure │
└────────────────┬────────────────┘
▼
┌─────────────────────────────────┐
│ 3. Visualize │
│ Spaghetti plot, look first │
└────────────────┬────────────────┘
▼
┌─────────────────────────────────┐
│ 4. Fit Models │
│ Baseline + growth models │
└────────────────┬────────────────┘
▼
┌─────────────────────────────────┐
│ 5. Compare Models │
│ LRT, AIC/BIC │
└────────────────┬────────────────┘
▼
┌─────────────────────────────────┐
│ 6. Check Diagnostics │
│ Fit indices, residuals │
└────────────────┬────────────────┘
▼
┌─────────────────────────────────┐
│ 7. Interpret Results │
│ Parameters, effect sizes │
└─────────────────────────────────┘
Step 1: Setup
# Load packages
library(tidyverse) # Data manipulation and plotting
library(lavaan) # SEM and growth models
library(MASS) # For mvrnorm (simulation)
# Set seed for reproducibility
set.seed(2024)
Step 2: Simulate Data
We'll create data with known population parameters so we can verify our model recovers them:
| Parameter | True Value |
|---|---|
| Intercept mean | 50 |
| Slope mean | 2 |
| Intercept variance | 100 (SD = 10) |
| Slope variance | 1 (SD = 1) |
| I-S covariance | -2 (r ≈ -0.20) |
| Residual variance | 25 (SD = 5) |
# Sample size
n <- 400
# Latent factor covariance matrix
psi <- matrix(c(100, -2,
-2, 1), nrow = 2)
# Generate individual intercepts and slopes
factors <- mvrnorm(n = n,
mu = c(50, 2),
Sigma = psi)
# Generate observed data
data_wide <- tibble(id = 1:n) %>%
mutate(
int_i = factors[, 1],
slp_i = factors[, 2],
y1 = int_i + slp_i * 0 + rnorm(n, 0, 5),
y2 = int_i + slp_i * 1 + rnorm(n, 0, 5),
y3 = int_i + slp_i * 2 + rnorm(n, 0, 5),
y4 = int_i + slp_i * 3 + rnorm(n, 0, 5),
y5 = int_i + slp_i * 4 + rnorm(n, 0, 5)
) %>%
select(id, y1:y5)
# Check structure
head(data_wide)
✅ Checkpoint: You should see a tibble with 6 rows and columns id, y1–y5. Values will differ due to random generation, but should be in the 30–70 range.
Step 3: Visualize
Always plot before modeling.
# Reshape for plotting
data_long <- data_wide %>%
pivot_longer(y1:y5, names_to = "wave", values_to = "y") %>%
mutate(time = as.numeric(gsub("y", "", wave)) - 1)
# Spaghetti plot
ggplot(data_long, aes(x = time, y = y, group = id)) +
geom_line(alpha = 0.15, color = "gray40") +
scale_x_continuous(breaks = 0:4, labels = paste("Wave", 1:5)) +
labs(x = "Time", y = "Score",
title = "Individual Growth Trajectories",
subtitle = "N = 400 participants, 5 waves") +
theme_minimal()
What to look for:
- General trend (up, down, flat?)
- Spread at baseline (intercept variance)
- Fan pattern (slope variance)
- Nonlinearity (curves vs. straight lines)
# Add mean trajectory
mean_traj <- data_long %>%
group_by(time) %>%
summarise(mean_y = mean(y))
ggplot(data_long, aes(x = time, y = y)) +
geom_line(aes(group = id), alpha = 0.1, color = "gray40") +
geom_line(data = mean_traj, aes(y = mean_y),
color = "steelblue", linewidth = 1.5) +
# Note: ggplot2 < 3.4 used 'size' instead of 'linewidth'
geom_point(data = mean_traj, aes(y = mean_y),
color = "steelblue", size = 3) +
scale_x_continuous(breaks = 0:4, labels = paste("Wave", 1:5)) +
labs(x = "Time", y = "Score",
title = "Individual Trajectories with Mean Overlay") +
theme_minimal()
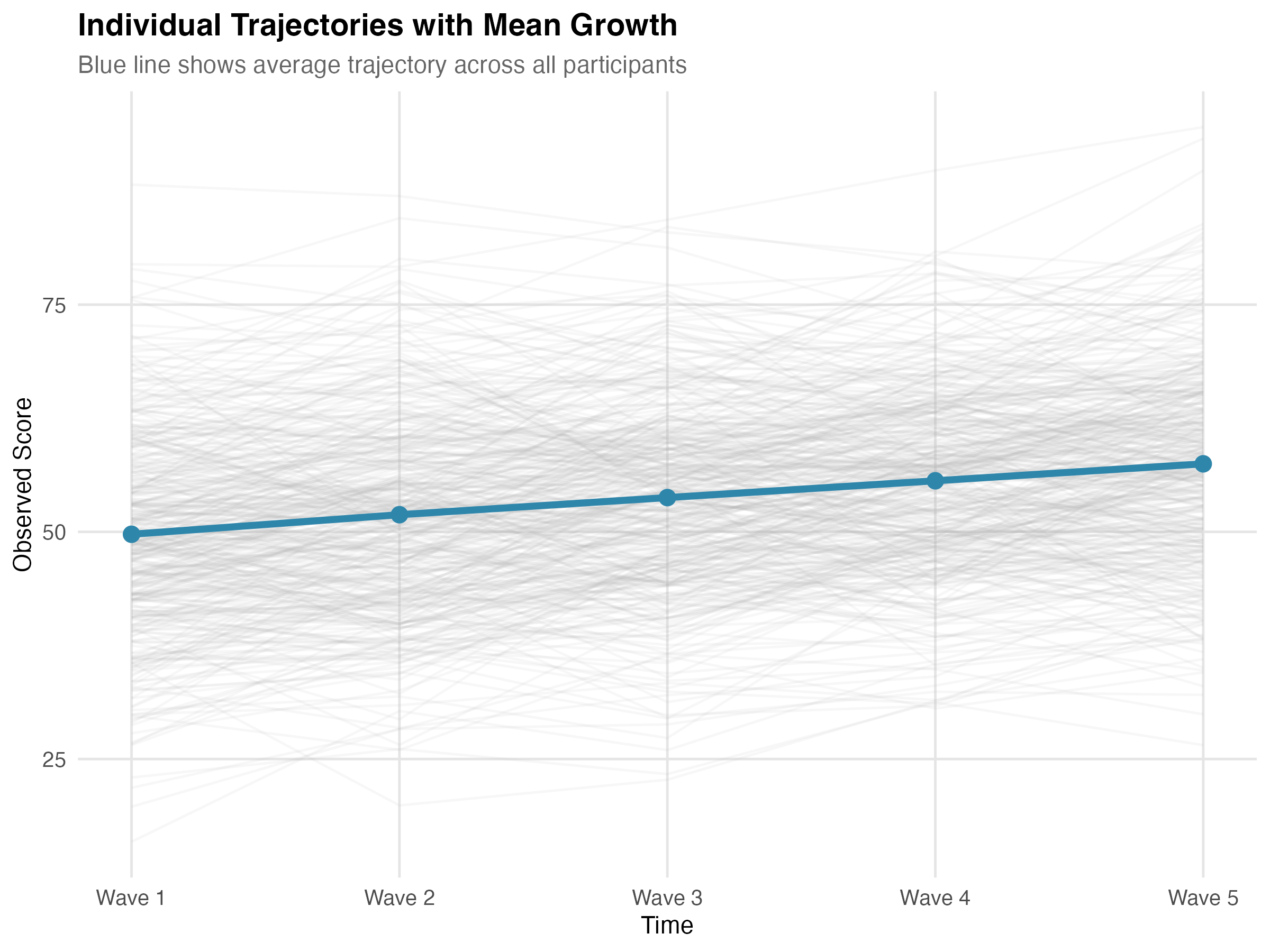
Figure: Individual trajectories (gray) with mean trajectory overlay (blue). The upward trend shows positive average growth; the spread shows individual differences.
✅ Checkpoint: Your plot should show upward-trending lines with visible spread at baseline and a "fan" pattern over time.
Step 4: Fit Models
No-Growth Model (Baseline)
Fit a constrained linear model with no growth—slope mean and variance fixed to zero:
model_nogrowth <- '
intercept =~ 1*y1 + 1*y2 + 1*y3 + 1*y4 + 1*y5
slope =~ 0*y1 + 1*y2 + 2*y3 + 3*y4 + 4*y5
# Constrain slope to "no growth"
slope ~ 0*1 # mean(slope) = 0
slope ~~ 0*slope # var(slope) = 0
intercept ~~ 0*slope # cov(i, s) = 0
'
fit_nogrowth <- growth(model_nogrowth, data = data_wide,
missing = "fiml")
Note: This model is nested within the linear growth model, allowing a valid likelihood ratio test.
Linear Growth Model
Add the slope factor:
model_linear <- '
intercept =~ 1*y1 + 1*y2 + 1*y3 + 1*y4 + 1*y5
slope =~ 0*y1 + 1*y2 + 2*y3 + 3*y4 + 4*y5
'
fit_linear <- growth(model_linear, data = data_wide,
missing = "fiml")
Note: We specify missing = "fiml" explicitly—good practice even when data are complete. For real data with potential non-normality, add estimator = "MLR" for robust standard errors.
Linear Growth with Equal Residuals
Test whether residual variances can be constrained equal:
model_linear_eq <- '
intercept =~ 1*y1 + 1*y2 + 1*y3 + 1*y4 + 1*y5
slope =~ 0*y1 + 1*y2 + 2*y3 + 3*y4 + 4*y5
# Constrain residual variances to equality
y1 ~~ rv*y1
y2 ~~ rv*y2
y3 ~~ rv*y3
y4 ~~ rv*y4
y5 ~~ rv*y5
'
fit_linear_eq <- growth(model_linear_eq, data = data_wide,
missing = "fiml")
Step 5: Compare Models
Is there growth?
anova(fit_nogrowth, fit_linear)
✅ Confirm: Δχ² should be significant (p < .001). This means linear growth improves fit over a flat trajectory (no-growth model).
Are equal residual variances justified?
anova(fit_linear_eq, fit_linear)
Use anova(constrained, unconstrained) so the Δχ² tests the relaxation of constraints.
✅ Confirm: If p > .05, equality is not rejected; prefer the simpler constrained model.
Information criteria
data.frame(
Model = c("No-growth", "Linear", "Linear (equal resid)"),
AIC = c(AIC(fit_nogrowth), AIC(fit_linear), AIC(fit_linear_eq)),
BIC = c(BIC(fit_nogrowth), BIC(fit_linear), BIC(fit_linear_eq))
) %>%
mutate(across(where(is.numeric), \(x) round(x, 1)))
# Note: R < 4.1 use function(x) instead of \(x)
✅ Confirm: Lower AIC/BIC = better fit. Linear models should beat no-growth.
Step 6: Check Diagnostics
Before interpreting results, verify model fit and check for problems.
Fit Indices
fitmeasures(fit_linear, c("chisq", "df", "pvalue", "cfi", "rmsea", "srmr"))
✅ Check fit indices:
- CFI > .95 ✓
- RMSEA < .08 ✓
- SRMR < .08 ✓
With small df, RMSEA can be unstable; interpret alongside CFI and SRMR.
See Reference for threshold details.
Modification Indices
modindices(fit_linear, sort = TRUE, minimum.value = 10)
What to look for:
- Residual covariances (e.g.,
y2 ~~ y3): May indicate autocorrelation - Large values (>10) suggest localized misfit
- Cross-loadings usually shouldn't be freed in LGCM
Consider multiplicity and substantive plausibility; avoid chasing small local improvements.
✅ Checkpoint: Few or no modification indices >10 indicates good specification.
Residual Correlations
resid(fit_linear, type = "cor")$cor %>% round(3)
Values should be near zero (|r| < 0.10). Large residual correlations mean the model doesn't fully capture those relationships. Thresholds like |r| < .10 are sample-size dependent; in large N, also inspect SEs/tests of residuals.
Check for Problematic Estimates
parameterEstimates(fit_linear) %>%
filter(op == "~~") %>%
select(lhs, rhs, est, se) %>%
mutate(flag = ifelse(est < 0, "NEGATIVE", ""))
Red flags:
- Negative variances (Heywood cases): Model misspecified or sample too small
- Very large SEs (SE > estimate): Parameter poorly identified
- Variances at exactly zero: May need constraints
Heywood case remedies:
- Try
estimator = "MLR"and/or rescale time to reduce collinearity - Temporarily constrain problematic residual variances to a small positive lower bound while diagnosing
✅ Confirm: All variance estimates should be positive. If any are negative, see Troubleshooting.
Step 7: Interpret Results
summary(fit_linear, fit.measures = TRUE, standardized = TRUE)
Compare Estimates to True Values
| Parameter | Estimate | SE | True Value |
|---|---|---|---|
| Intercept mean | 49.82 | 0.51 | 50 |
| Slope mean | 2.04 | 0.06 | 2 |
| Intercept variance | 98.34 | 8.12 | 100 |
| Slope variance | 0.97 | 0.12 | 1 |
| I-S covariance | -1.89 | 0.62 | -2 |
| Residual variance | 24.56 | 1.23 | 25 |
Values shown are illustrative from one simulated run; your numbers will differ with seed, RNG, and package versions (even with the same seed).
✅ Success: Estimates closely recover the true population parameters. This confirms the model is working correctly.
Interpret Each Parameter
- Intercept mean ≈ 50: Average score at Wave 1
- Slope mean ≈ 2: Average increase per wave
- Intercept variance ≈ 100 (SD ≈ 10): People differed in starting levels
- Slope variance ≈ 1 (SD ≈ 1): People differed in growth rates
- I-S covariance ≈ -2: Higher starters grew slightly slower (see correlation below)
Intercept-Slope Correlation
Convert the covariance to a correlation for interpretability:
cov_is <- -1.89
var_i <- 98.34
var_s <- 0.97
r_is <- cov_is / sqrt(var_i * var_s)
r_is # ≈ -0.19
The correlation of -0.19 is small—higher starters grew slower, but most still improved.
Proportion with Positive Slopes
A positive slope mean doesn't mean everyone improved. Check:
slope_mean <- 2.04
slope_sd <- sqrt(0.97) # ≈ 0.98
# P(slope > 0) — theoretical
pnorm(0, mean = slope_mean, sd = slope_sd, lower.tail = FALSE)
# ≈ 0.98
# Empirical check using factor scores (posterior means)
fs <- lavPredict(fit_linear, type = "lv")
mean(fs[, "slope"] > 0)
# Should be close to the theoretical value
✅ Checkpoint: ~98% of participants had positive slopes. With slope mean = 2 and SD ≈ 1, almost everyone improved. The empirical proportion from factor scores should match the theoretical pnorm() result.
Variance Explained
inspect(fit_linear, "r2")
Expected R² values: 0.80–0.89 across waves.
Interpretation: The trajectory (intercept + slope) explains 80–89% of variance at each wave. The remaining 10–20% is residual variance (measurement error, occasion-specific factors).
✅ Check: R² values of 0.70–0.90 are typical for well-fitting LGCMs.
Written Summary
We estimated a linear latent growth model for 400 participants across 5 waves. The model fit well (χ²(10) = 12.35, p = .26; CFI = 0.998; RMSEA = 0.024; SRMR = 0.025).
On average, participants started at 49.82 (SE = 0.51) and increased by 2.04 units per wave (SE = 0.06), both p < .001. Significant individual differences emerged in starting levels (variance = 98.34, SD ≈ 10) and growth rates (variance = 0.97, SD ≈ 1). The negative intercept-slope covariance (-1.89, p = .002, r ≈ -0.19) indicates that participants who started higher grew slightly slower.
Complete Script
# ============================================
# LGCM Complete Worked Example
# ============================================
library(tidyverse)
library(lavaan)
library(MASS)
set.seed(2024)
# Simulate data
n <- 400
psi <- matrix(c(100, -2, -2, 1), nrow = 2)
factors <- mvrnorm(n, mu = c(50, 2), Sigma = psi)
data_wide <- tibble(id = 1:n) %>%
mutate(
int = factors[,1], slp = factors[,2],
y1 = int + slp*0 + rnorm(n, 0, 5),
y2 = int + slp*1 + rnorm(n, 0, 5),
y3 = int + slp*2 + rnorm(n, 0, 5),
y4 = int + slp*3 + rnorm(n, 0, 5),
y5 = int + slp*4 + rnorm(n, 0, 5)
) %>% select(id, y1:y5)
# Visualize
data_long <- data_wide %>%
pivot_longer(y1:y5, names_to = "wave", values_to = "y") %>%
mutate(time = as.numeric(gsub("y", "", wave)) - 1)
ggplot(data_long, aes(x = time, y = y, group = id)) +
geom_line(alpha = 0.15) +
theme_minimal() +
labs(title = "Individual Trajectories")
# Fit models
model_nogrowth <- '
intercept =~ 1*y1 + 1*y2 + 1*y3 + 1*y4 + 1*y5
slope =~ 0*y1 + 1*y2 + 2*y3 + 3*y4 + 4*y5
slope ~ 0*1; slope ~~ 0*slope; intercept ~~ 0*slope
'
model_lin <- '
intercept =~ 1*y1 + 1*y2 + 1*y3 + 1*y4 + 1*y5
slope =~ 0*y1 + 1*y2 + 2*y3 + 3*y4 + 4*y5
'
fit_nogrowth <- growth(model_nogrowth, data = data_wide, missing = "fiml")
fit_lin <- growth(model_lin, data = data_wide, missing = "fiml")
# Compare and summarize
anova(fit_nogrowth, fit_lin)
summary(fit_lin, fit.measures = TRUE, standardized = TRUE)
fitmeasures(fit_lin, c("chisq", "df", "pvalue", "cfi", "rmsea", "srmr"))
inspect(fit_lin, "r2")
Adapt to Your Data
To use this workflow with your own dataset:
1. Read and Prepare Data
library(readr)
library(dplyr)
# Read your data (wide format: one row per person)
data_wide <- read_csv("my_longitudinal_data.csv") %>%
select(id, outcome_t1, outcome_t2, outcome_t3, outcome_t4, outcome_t5)
2. Update Variable Names
Replace y1–y5 with your variable names:
model <- '
intercept =~ 1*outcome_t1 + 1*outcome_t2 + 1*outcome_t3 + 1*outcome_t4 + 1*outcome_t5
slope =~ 0*outcome_t1 + 1*outcome_t2 + 2*outcome_t3 + 3*outcome_t4 + 4*outcome_t5
'
fit <- growth(model, data = data_wide, missing = "fiml")
summary(fit, fit.measures = TRUE, standardized = TRUE)
3. Adjust Time Coding (if needed)
If waves are unequally spaced, adjust slope loadings to reflect actual time. For example, measurements at baseline, 1, 3, 6, and 12 months:
model <- '
intercept =~ 1*y_0mo + 1*y_1mo + 1*y_3mo + 1*y_6mo + 1*y_12mo
slope =~ 0*y_0mo + 1*y_1mo + 3*y_3mo + 6*y_6mo + 12*y_12mo
'
# Now slope = change per month
Rescaling time (e.g., months → years) rescales the slope mean and variance (per-unit); choose units that are meaningful for interpretation.
✅ Checkpoint: Once your basic model runs, layer on diagnostics, model comparisons, and predictors exactly as shown above.
Next Steps
- Need syntax or thresholds? → Reference
- Hit an error? → Reference: Troubleshooting
- Back to overview → LGCM Guide